Large research networks, the implementation of clinical-epidemiological studies, but also the establishment of registries and cohorts, require data processing that complies with data protection regulations. In accordance with Art. 32 para. 1a GDPR, the use of pseudonyms helps to ensure an appropriate level of data processing protection.
Large research networks, the implementation of clinical-epidemiological studies, but also the establishment of registries and cohorts, require data processing that complies with data protection regulations. In accordance with Art. 32 para. 1a GDPR, the use of pseudonyms helps to ensure an appropriate level of data processing protection.
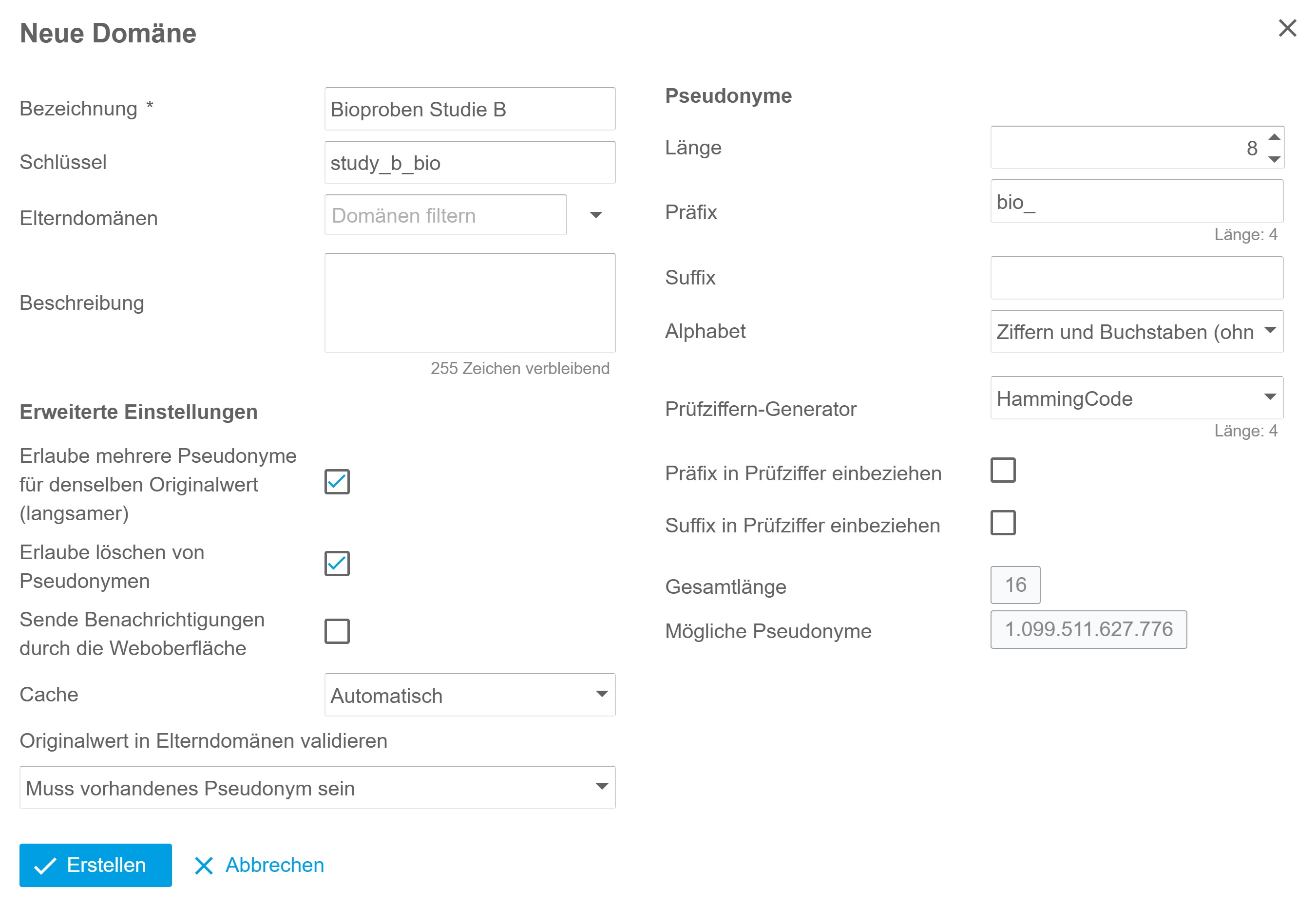
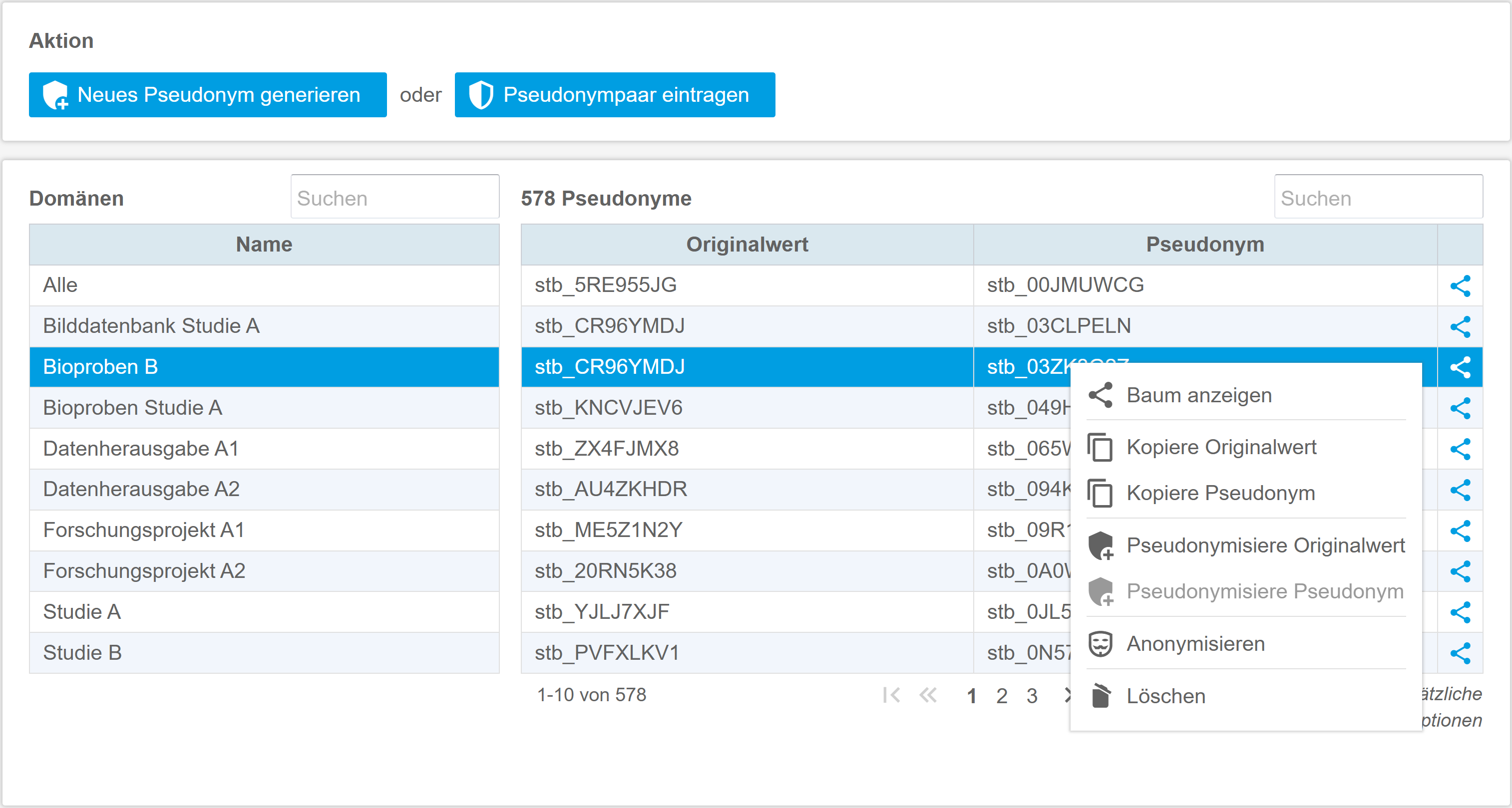
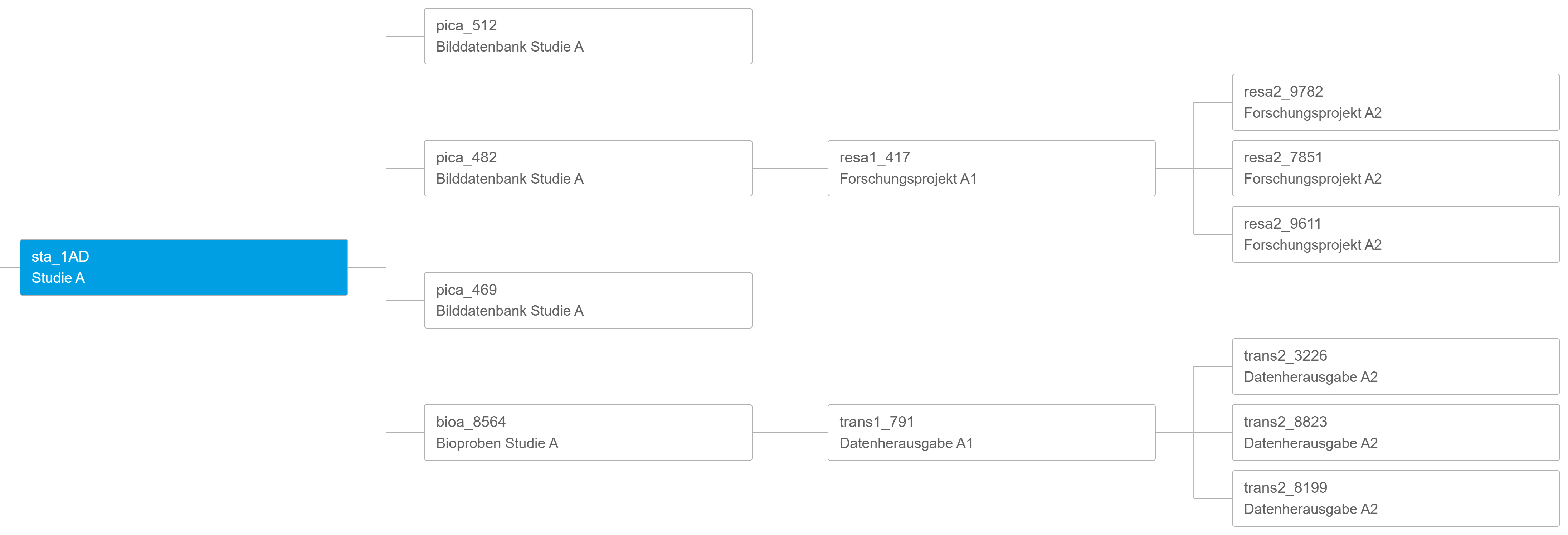
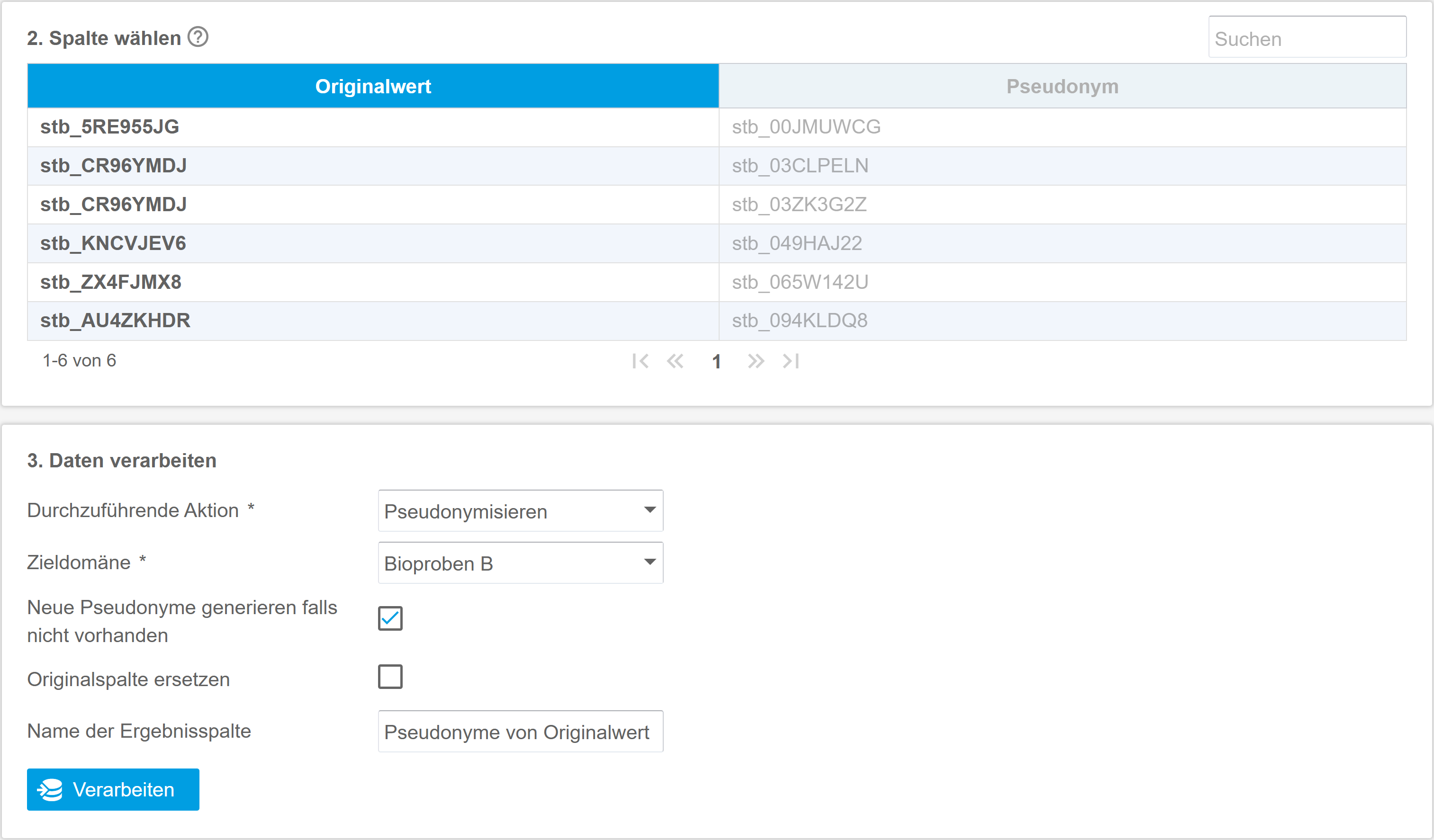
The gPAS® was developed for this specific purpose. It is used to generate and manage pseudonyms. The domain concept enables the structured management of pseudonyms per data source, application context (data collection, data transfer) or location. The characters (alphabet) and algorithms used can be freely defined. The linking of domains enables the creation of hierarchies and the generation of pseudonyms of any level (pseudonymisation of pseudonyms). The intuitive web interface supports the creation and resolving of pseudonyms. If necessary, pseudonyms can be created temporarily or anonymised in order to take account of the ‘right to be forgotten’ of data subjects. At the same time, gPAS also supports large amounts of data by means of list processing. The dashboard provides an overview of the figures for generated pseudonyms and configured domains. Users can get started quickly and easily with the manual provided, which guides them step by step through the functionalities and necessary configurations.
Download
Are you interested in the gPAS? You can download it here and simply start it using Docker. Alternatively, you can try out the gPAS in our live demo.
Interfaces
Documentation
User projects
The solutions of the Trusted Third Party of the University Medicine Greifswald are becoming increasingly widespread in the community. More and more consortia, sites and projects have made a conscious decision to use our solutions to realise their individual application scenarios. Users include registries and cancer registries, local trust centres of the MII and the NUM, various study projects and selected DZGs.
We have created the THS Community Dialogue to exchange experiences and share results with each other.
Funding and publications
The gPAS® is being developed by the University Medicine Greifswald (Institute for Community Medicine) and was published in 2013 as part of the MOSAIC project (funded by the DFG HO 1937/2-1). Selected functions of the gPAS® were implemented as part of the MIRACUM project (funded by the BMBF 01ZZ1801M). The gPAS® is continuously being further enhanced on the basis of project-specific requirements and feedback from the user community.
Selected publications
- MOSAIC – A Modular Approach to Data Management in Epidemiological Studies
- A workflow-driven approach to integrate generic software modules in a Trusted Third Party
- Integration of trusted third party software into an EDC system for data protection-compliant identity management, consent management and pseudonymization in medical research studies
- Pseudonymisierung in REDCap mit E-PIX und gPAS